Epigenetic: A fine interplay exists between sensory experience and innate genetic programs leading to the sculpting of neuronal circuits during early brain development. Recent evidence suggests that the dynamic regulation of gene expression through epigenetic mechanisms is at the interface between environmental stimuli and long lasting molecular, cellular and complex behavioral phenotypes acquired during periods of developmental plasticity. Understanding these mechanisms may give insight into the formation of critical periods and provide new strategies for increasing plasticity and adaptive change in adulthood.
Introduction
During early development, neuronal circuits are created and connections between neurons undergo remodeling as they develop their adult functional properties in response to the surrounding environment. The adult brain loses this extraordinary plasticity. Recent findings support a key role of epigenetic factors in mediating the effects of sensory experience on site-specific gene expression, synaptic transmission, and behavioral phenotypes. Here we review recent evidence implicating multiple epigenetic mechanisms in experience-dependent changes during development and discuss their role in critical period expression in the developing and adult brain.Epigenetics: Molecular Mechanisms Of Gene Regulation
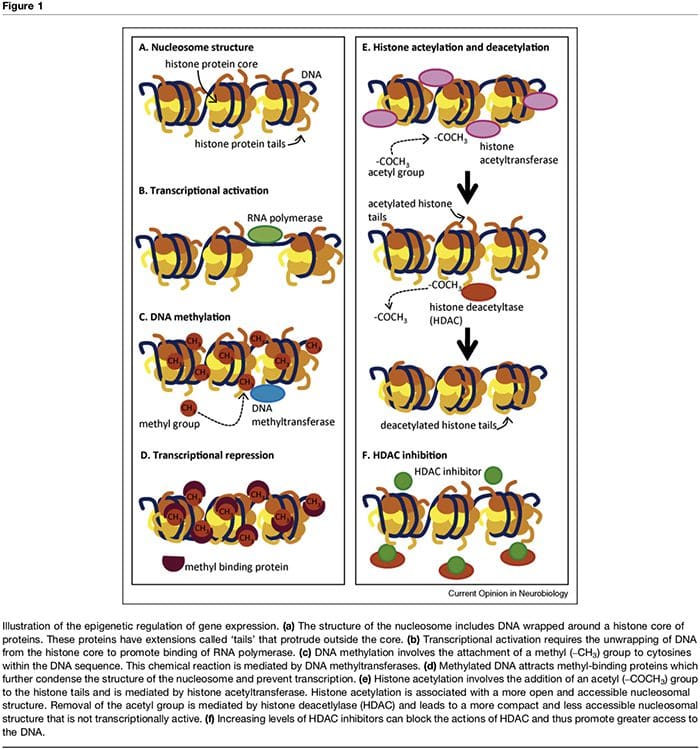
The term ‘epigenetic’ refers to chromatin modifications which alter gene expression without affecting DNA sequence. The factors that contribute to the epigenetic regulation of transcriptional activity are numerous and include microRNA [1], DNA methylation [2,3] and post- translational modifications of nucleosomal histones [2,4]. DNA methylation refers to a chemical modification to DNA whereby cytosine is converted to 5-methylcytosine with the consequence of reduced accessibility of the DNA to transcription factors (Figure 1a–d). These modi- fications can be stable and heritable and provide a critical mechanism in cellular differentiation [3]. The process of methylation is dependent on the presence of methyl donors (provided by nutrients such as folic acid, meth- ionine and choline) and methyltransferases which med- iate either maintenance (i.e. DNMT1) or de novo DNA methylation (i.e. DNMT3). Transcriptional repression associated with DNA methylation is further sustained through methyl-binding proteins such as MeCP2 [5]. Epigenetic control of gene expression is also mediated through multiple post-translational modifications of histone proteins, including methylation, acetylation and ubiquination, which can alter the accessibility of DNA and the density of chromatin structure (Figure 1e,f). In particular, histone acetylation is associated with increased transcriptional activity whereas histone deacetylation is associated with transcriptional repression. The acetylation state of these nucleosomal proteins is controlled by the presence of histone acetyltransferases (HATs), histone deacetylases (HDACs), which are recruited by methyl-binding proteins, and by HDAC inhibitors, which effectively increase gene expression through shifting histones to an acetylated state [2,6]. The timing and degree of gene expression are controlled through these complex mechanisms, thus providing a link between single genotypes and multiple phenotypes.Epigenetic Factors & The Influence Of Early Life Experiences
In mammalian development, the prenatal and postnatal periods are characterized by rapid changes in neuronal organization, thus providing a critical window of opportunity during which environmental experiences can lead to long-term influences on brain and behavior. There is increasing evidence for the role of epigenetic factors in mediating the relationship between these experiences and long-term outcomes. Mueller and Bale [7] have recently demonstrated decreased DNA methylation of the corticotrophin-releasing-factor (CRF) gene promotor and increased methylation of the glucocorticoid receptor (GR) exon 17 promotor region in hypothalamic tissue of adult male mice born to gestationally stressed females. These epigenetic modifications are associated with exposure to stress during the early stages of prenatal development and may involve dysregulation of placental gene expression. The nutritional environment during fetal development has likewise been demonstrated to influence growth, metabolism and brain development and there is increasing evidence that dietary levels of methyl-donors can epigenetically alter gene expression in offspring [8,9]. In rats, Lillycrop et al. [10] illustrate that GR 110 and PPARa (peroxisome proliferator-activated receptor alpha) gene promotor methylation is reduced in the hepatic tissue of offspring born to protein restricted dams whereas methylation is increased in offspring of dams whose diet is supplemented with methyl donors [10,11]. These effects may be related to DNMT1 expression, which is likewise decreased with dietary protein restriction [11]. Prenatal nutritional regulation of DNA methylation has similarly been observed in brain tissue associated with levels of DNMT1 expression [12], suggesting that in the rapid period of cell division ocurring during fetal development, the level of methyl donors can have a significant impact on transcriptional activity that is maintained into adulthood.The role of epigenetic modification in sustaining the effects of environmental experience has also been demonstrated in the context of postnatal mother–infant interactions. Individual variations in maternal care during the immediate postpartum period in rats are associated with changes in offspring hypothalamic-pituitary-adrenal (HPA) activity, neuroendocrine systems involved in reproduction and hippocampal plasticity [13]. Analyses of levels of promotor methylation within the hippocampal GR 17 and hypothalamic ERa genes in offspring of rat dams that provide high vs. low levels of maternal care indicate that high levels of care are associated with decreased promotor methylation and thus increased gene expression [14,15]. Though the route through which these epigenetic changes are mediated is not yet clear, there is evidence for increased binding of nerve growth factor-inducible protein A (NGFI-A) to the GR exon 17 promoter amongst offspring who receive high levels of care in infancy [15] and in vitro models suggest that NGFI-A up-regulation is associated with histone acetylation, DNA demethylation, and activation of the exon 17 GR promoter [16]. The relevance of these effects in humans has recently been demonstrated by Oberlander et al. [17] in the analysis of methylation status of the GR promotor at NGFI-A binding sites in cord blood mononuclear cells of infants exposed to third trimester maternal depressed or anxious mood. Maternal depression was found to be associated with increased GR 1F promotor methylation in fetal blood samples and these methylation patterns predicted HPA reactivity in infants at 3 months of age [17]. Analysis of hippocampal tissue from suicide victims with a history of childhood abuse similarly indicates lower GR expression and higher GR 1F promotor methylation associated with disruptions of the early environment and confirms the findings from rodent studies that differential NGFI-A binding is a functional consequence of these epigenetic effects [18]. However, the impact of perinatal mother– infant interactions is not limited to GR regulation as illustrated by Roth et al. [19] examining the effects of postnatal abuse on offspring brain derived neurotrophic factor (BDNF) methylation [19]. In rats, an increase in methylation of exon IV of the BDNF promotor and consequent decrease in BDNF mRNA in the prefrontal cortex was found in association with exposure to periods of abusive maternal care (dragging, rough handling, etc.). As was the case with the effects of individual differences in maternal care, these effects emerged in infancy and were sustained into adulthood. Moreover, these effects on BDNF exon IV methylation are perpetuated to the F1 generation suggesting a role for epigenetic mechanisms in transgenerational effects [20].
Development Across The Lifespan: Epigenetics & Experience Dependent Plasticity
The previous section highlights the stable effects of early life experiences and how these events become encoded at a molecular level. Another approach to the study of epigenetics and development comes from studies of synaptic plasticity during the expression of long-term potentiation (LTP) and memory consolidation. High levels of maternal care and exposure to juvenile environ- mental enrichment (EE) have been demonstrated to improve capacity for learning and memory associated with LTP enhancement [21,22]. Moreover, recent evidence suggests that EE modulates NMDAr/p38/LTP signaling pathways in the hippocampus and improves contextual fear memory formation across generations such that offspring of enriched mothers likewise show enhanced LTP even when cross-fostered at birth to non- enriched mothers [23]. Environmental enrichment has been associated with increased histone acetylation in the hippocampus and improved spatial memory [24,25]. Pharmacological targeting of the epigenome has been used to demonstrate the role of histone acetylation and DNA methylation in the consolidation of long-term memory [26]. Treatment with zebularine (an inhibitor or DNA methyltransferases) has been shown to block memory formation and reduce histone acetylation following con- textual fear conditioning in adult rats [27] whereas treatment with the HDAC inhibitor sodium butyrate lead to enhanced formation of contextual fear memories [28]. The particular HDAC target of these inhibitors may be HDAC2 as recent evidence has emerged illustrating decreased synaptic plasticity and memory formation in mice over-expressing HDAC2 but not HDAC1; with the converse effect in HDAC2-deficient mice [29]. These studies illustrate a possible relationship between synaptic activity and histone acetylation/DNA methylation in mature neurons, suggesting that there is continued plasticity in these epigenetic systems beyond the prenatal and postnatal periods of development.Epigenetic Mechanism & The Regulation Of Synaptic Transmission
Activity-dependent changes in gene expression within neuronal pathways during development may serve as a critical pathway linking experience of the external environment and epigenetic modifications within the cell nucleus. In a recent study, Monteggia and colleagues elegantly demonstrated that spontaneous synaptic trans- mission in hippocampal neurons is regulated by alterations in DNA methylation that occur in response to synaptic activity [30]. Treatment with a DNMT inhibitor lead to a significant decrease in frequency of miniature excitatory post-synaptic currents (mEPSCs) and rate of spontaneous synaptic vesicle fusion correlated with a decrease in BDNF promoter I methylation and increased BDNF expression. This effect was blocked with inhibition of synaptic activity and reductions in mEPSCs were prevented in the absence of MeCP2. These results strongly suggest a role for DNA methylation/MeCP2 pathways in the control of synaptic function. Activity- dependent phosphorylation of MeCP2 via Ca2+-calmodulindependent kinase II has been shown to cause MeCP2 dissociation from target genes and relieve transcriptional repression [31]. Consequently, genes such as BDNF are increased in expression leading to normal dendritic patterning and dendritic spine development [32]. These findings suggest an epigenetic mechanism through which neurons can monitor alterations in activity level and adjust neurotransmitter output via altered gene expression with consequences for network excitability and circuit refinement. Impairments in these MeCP2 pathways may lead to several neurodevelopmental abnormalities including Rett syndrome, infantile autism, mental retardation, and schizophrenia [33] and targeted deletion of MeCP2 in the amygdala has recently been demonstrated to impair learning and memory and lead to increased anxiety-like behavior in mice [34].Epigenetic Control Of Critical Period Plasticity
Though epigenetic mechanisms have certainly been implicated in mediating the high levels of plasticity in early development, it is also possible to view the decreased plasticity and sensitivity that occurs later in development from an epigenetic perspective. Neocortical circuits are extremely sensitive to manipulations of the sensory environment during restricted temporal windows of postnatal development called ‘critical periods’. For example, an imbalance in binocular vision during child- hood affects perception leading to amblyopia or ‘lazy eye’. Monocular deprivation (MD) reproduces this classical paradigm of experience-dependent plasticity [35]. The striking physiological effect of MD is a shift in visual cortical neuron response in favor of the non-deprived eye; an example of ocular dominance (OD) plasticity. The critical period during which this OD plasticity occurs is defined by the activation and subsequent inhibition of specific molecular pathways involving signaling molecules such as aCaMKII, calcineurin, PKA, ERK, and CREB [36]. Recently, Pizzorusso and colleagues identified rapid increases in ERK-dependent phosphorylation of histones associated with activation of the juvenile visual cortex and a developmental downregulation of this effect in older mice [37]. In adult mice, the reduced OD plasticity can be reinstated through treatment with the HDAC inhibitor trichostatin A (TSA). Multiple cellular mechanisms might contribute to experience-dependent plasticity expression [38]. Further work is necessary to understand if epigenetic mechanisms are generally acting in all cellular substrates or only within a specific subset.Myelin maturation has also been proposed as one of the major factors contributing to decreased neuronal plasticity. During the onset of critical period plasticity, oligodendrocytes start to express specific myelin structural proteins, including myelin basic protein (MBP), myelin-associated glycoprotein (MAG), myelin oligodendrocyte glycoprotein (OMgp) and myelin-associated oligodendrocyte basic protein (MOBP) [39]. As myelination reaches adult levels, OD plasticity is strongly reduced or absent. MAG and OMgp may contribute to critical period closure through activation of Nogo receptors. Indeed, mice lacking Nogo receptors exhibit OD plasticity even in adulthood [40]. Manipulation of epigenetic status of oligodendrocytes may also be an effective strategy for modulating plasticity. Casaccia-Bonnefil and colleagues have shown that histone modifications are involved in oligodendrocyte precursor cell (OPC) differentiation during development and in recovery from injury [41– 43]. Administration of the HDAC inhibitor valproic acid during the critical period of myelination onset was found to prevent the OPC maturation into myelinating cells. These results suggest that HDAC activity during a specific temporal window of postnatal development is required for OPC differentiation and myelination. At later developmental stages, histone deacetylation subsides and is replaced by repressive histone methylation and the establishment of a compact chromatin structure, characteristic of the differentiated oligodendrocyte phenotype [43]. Shen et al. [44] found that in response to damage of oligodendrocytes, robust remyelination occurred in juvenile but not in older animals with the new myelin synthesis preceded by down regulation of oligodendrocyte differentiation inhibitors and neural stem cell markers and the recruitment of HDACs to promoter regions. This HDAC recruitment is inefficient in older brains, allowing for the accumulation of transcriptional inhibitors and prevention of myelin gene expression. This age-depend- ent effect can be induced in young mice treated with HDAC inhibitors during the period when damage to oligodentrocytes is occurring. Thus, there are epigenetic changes that are characteristic of periods of developmental plasticity that could provide a target for therapeutic intervention in the event of CNS damage. The use of HDAC inhibitors to increase plasticity in the brain may be a promising therapeutic approach as there is conver- ging evidence from rodent models that treatment with these compounds (1) can lead to dramatic shifts in gene expression and behavior in adult offspring who have received low levels of maternal care [15] and (2) mimic the effects of EE on reversal of neurodevelopmental abnormalities [24]. Rather than producing a generalized increase in transcription, these compounds lead to acti- vation of a specific subset of genes [45–47], suggesting possible targeted intervention to reinstate plasticity in adult brain.
Conclusions
There is converging evidence for the role of epigenetic modifications such as histone acetylation and DNA meth- ylation in both the stability and plasticity of developing neuronal circuits. The persistent effects on gene expression that can be achieved through these mechanisms provide a biological route through which environmental experiences can become embedded, leading to long-term changes in neurobiology and behavior. Enhancing plasticity in the adult brain is an exciting prospect and there is certainly evidence emerging that suggest the possible use of epigenetic factors to induce a ‘younger’ brain. The challenge of future studies is to establish the pathways through which site-specific and gene-specific transcriptional modifications can be achieved and to better understand the route through which experiences across the lifespan induce this molecular plasticity.Michela Fagiolini 1, Catherine L Jensen 2 and Frances A Champagne 2
Current Opinion in Neurobiology 2009, 19:1–6This review comes from a themed issue on Development
Edited by Takao Hensch and Andrea Brand
0959-4388/$ – see front matter Published by Elsevier Ltd.
DOI 10.1016/j.conb.2009.05.009
Corresponding author: Champagne, Frances A (fac2105@columbia.edu)
References:
1. Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF: RNA
regulation of epigenetic processes. Bioessays 2009, 31:51-59.
2. Feng J, Fouse S, Fan G: Epigenetic regulation of neural gene
expression and neuronal function. Pediatr Res 2007, 61:58R63R.
3. Razin A: CpG methylation, chromatin structure and gene
silencing-a three-way connection. EMBO J 1998, 17:4905-4908.
4. Fukuda S, Taga T: Cell fate determination regulated by a
transcriptional signal network in the developing mouse brain.
Anat Sci Int 2005, 80:12-18.
5. Fan G, Hutnick L: Methyl-CpG binding proteins in the nervous
system. Cell Res 2005, 15:255-261.
6. Strathdee G, Brown R: Aberrant DNA methylation in cancer:
potential clinical interventions. Expert Rev Mol Med 2002,
2002:1-17.
7. Mueller BR, Bale TL: Sex-specific programming of offspring
emotionality after stress early in pregnancy. J Neurosci 2008,
28:9055-9065.
Rodent study illustrating alterations in DNA methylation of placental and
brain tissue following exposure to gestational stress, providing a possible
mechanism mediating the long-term neurobiological effects of prenatal
exposure to elevated maternal HPA activity
8. Hoet JJ, Hanson MA: Intrauterine nutrition: its importance
during critical periods for cardiovascular and endocrine
development. J Physiol 1999, 514(Pt 3):617-627.
9. Zeisel SH: Importance of methyl donors during reproduction.
Am J Clin Nutr 2009, 89:673S-677S.
10. Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA,
Burdge GC: Feeding pregnant rats a protein-restricted diet
persistently alters the methylation of specific cytosines in the
hepatic PPAR alpha promoter of the offspring. Br J Nutr 2008,
100:278-282.
11. Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM,
Jackson AA, Burdge GC: Induction of altered epigenetic
regulation of the hepatic glucocorticoid receptor in the
offspring of rats fed a protein-restricted diet during pregnancy
suggests that reduced DNA methyltransferase-1 expression is
involved in impaired DNA methylation and changes in histone
modifications. Br J Nutr 2007, 97:1064-1073.
Illustrates the impact of methyl donors in the maternal diet on offspring
DNA methylation and histone acetylation patterns, providing an important
link between nutrition and gene regulation
12. Kovacheva VP, Mellott TJ, Davison JM, Wagner N, LopezCoviella
I, Schnitzler AC, Blusztajn JK: Gestational choline
deficiency causes global- and Igf2 gene- DNA
hypermethylation by upregulation of Dnmt1 expression. J Biol
Chem 2007, 282:31777-31788.
13. Meaney MJ: Maternal care, gene expression, and the
transmission of individual differences in stress reactivity
across generations. Annu Rev Neurosci 2001, 24:1161-1192.
14. Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M,
Meaney MJ: Maternal care associated with methylation of the
estrogen receptor-alpha1b promoter and estrogen receptoralpha
expression in the medial preoptic area of female
offspring. Endocrinology 2006, 147:2909-2915.
15. Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S,
Seckl JR, Dymov S, Szyf M, Meaney MJ: Epigenetic programming
by maternal behavior. Nat Neurosci 2004, 7:847-854.
16. Weaver IC, D’Alessio AC, Brown SE, Hellstrom IC, Dymov S,
Sharma S, Szyf M, Meaney MJ: The transcription factor nerve
growth factor-inducible protein a mediates epigenetic
programming: altering epigenetic marks by immediate-early
genes. J Neurosci 2007, 27:1756-1768.
17. Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S,
Devlin AM: Prenatal exposure to maternal depression,
neonatal methylation of human glucocorticoid receptor gene
(NR3C1) and infant cortisol stress responses. Epigenetics
2008, 3:97-106.
Provides evidence for the relevance of epigenetic mechanisms in mediating
the effects of maternal mood on infant development. Illustrates the
translation of experimental approaches conducted in animal models to
the study of clinically relevant issues in humans
18. McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B,
Szyf M, Turecki G, Meaney MJ: Epigenetic regulation of the
glucocorticoid receptor in human brain associates with
childhood abuse. Nat Neurosci 2009, 12:342-348.
Provides compelling evidence for the presence of differential methylation
of GR with consequences for gene expression in the human hippocampus
as a function of early childhood abuse using brain tissue obtained
from suicide victims
19. Roth TL, Lubin FD, Funk AJ, Sweatt JD: Lasting epigenetic
influence of early-life adversity on the BDNF gene. Biol
Psychiatry 2009, 65:760-769.
Rodent study of the transgenerational impact of exposure to maternal
abuse in infancy and the role of differential methylation of BDNF in the
prefrontal cortex in mediating these effects
20. Champagne FA: Epigenetic mechanisms and the
transgenerational effects of maternal care. Front
Neuroendocrinol 2008, 29:386-397.
21. Bruel-Jungerman E, Laroche S, Rampon C: New neurons in the
dentate gyrus are involved in the expression of enhanced
long-term memory following environmental enrichment.
Eur J Neurosci 2005, 21:513-521.
22. Champagne DL, Bagot RC, van Hasselt F, Ramakers G,
Meaney MJ, de Kloet ER, Joels M, Krugers H: Maternal care and
hippocampal plasticity: evidence for experience-dependent
structural plasticity, altered synaptic functioning, and
differential responsiveness to glucocorticoids and stress.
J Neurosci 2008, 28:6037-6045.
23. Arai JA, Li S, Hartley DM, Feig LA: Transgenerational rescue of a
genetic defect in long-term potentiation and memory
formation by juvenile enrichment. J Neurosci 2009, 29:1496-
1502.
Rodent study illustrating the transgenerational impact of environmental
enrichment on LTP suggesting that genetically induced deficits can be
overcome through environmental conditions experienced by a previous
generation
24. Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH: Recovery
of learning and memory is associated with chromatin
remodelling. Nature 2007, 447:178-182.
Rodent study showing that environmental enrichment increases histone
acetylation in the hippocampus. HDAC inhibitors induce increased spatial
memory in a neurodegenerative disorder mouse model
25. Williams BM, Luo Y, Ward C, Redd K, Gibson R, Kuczaj SA,
McCoy JG: Environmental enrichment: effects on spatial
memory and hippocampal CREB immunoreactivity. Physiol
Behav 2001, 73:649-658.
26. Sweatt JD: Experience-dependent epigenetic modifications in
the central nervous system. Biol Psychiatry 2009, 65:191-197.
27. Lubin FD, Roth TL, Sweatt JD: Epigenetic regulation of BDNF
gene transcription in the consolidation of fear memory.
J Neurosci 2008, 28:10576-10586.
Recent paper from a series of investigations by the Sweatt lab illustrating
the dynamic changes to DNA methylation which occur during the process
of learning and the critical role of these modifications in the consolidation
of memory
28. Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P,
Malone LM, Sweatt JD: Evidence that DNA (cytosine-5)
methyltransferase regulates synaptic plasticity in the
hippocampus. J Biol Chem 2006, 281:15763-15773.
29. Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N,
Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R et al.: HDAC2
negatively regulates memory formation and synaptic
plasticity. Nature 2009, 459:55-60.
Study in mice examining the particular HDAC target through which HDAC
inhibitors exert enhancements in synaptic plasticity and memory. Using
targeted up- and downregulation of HDAC2 the authors illustrate the
importance of levels of this enzyme in mediating cognitive enhancement
30. Nelson ED, Kavalali ET, Monteggia LM: Activity-dependent
suppression of miniature neurotransmission through the
regulation of DNA methylation. J Neurosci 2008, 28:395-406.
This paper focuses on the regulation of DNA methylation by NMDA
receptor-mediated synaptic activity within mature neurons and how
epigenetic alterations affect basal synaptic function. These findings
suggest a synaptic basis for neurological symptoms associated with
neurodevelopmental disorders such as Rett syndrome
31. Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC,
Jaenisch R, Greenberg ME: Derepression of BDNF transcription
involves calcium-dependent phosphorylation of MeCP2.
Science 2003, 302:885-889.
32. Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L,
Chen WG, Lin Y, Savner E, Griffith EC et al.: Brain-specific
phosphorylation of MeCP2 regulates activity-dependent Bdnf
transcription, dendritic growth, and spine maturation. Neuron
2006, 52:255-269.
33. Moretti P, Zoghbi HY: MeCP2 dysfunction in Rett syndrome and
related disorders. Curr Opin Genet Dev 2006, 16:276-281.
34. Adachi M, Autry AE, Covington HE 3rd, Monteggia LM: MeCP2-
mediated transcription repression in the basolateral amygdala
may underlie heightened anxiety in a mouse model of Rett
syndrome. J Neurosci 2009, 29:4218-4227.
35. Tropea D, Van Wart A, Sur M: Molecular mechanisms of
experience-dependent plasticity in visual cortex. Philos Trans
R Soc Lond B Biol Sci 2009, 364:341-355.
36. Medini P, Pizzorusso T: Visual experience and plasticity of the
visual cortex: a role for epigenetic mechanisms. Front Biosci
2008, 13:3000-3007.
37. Putignano E, Lonetti G, Cancedda L, Ratto G, Costa M, Maffei L,
Pizzorusso T: Developmental downregulation of histone
posttranslational modifications regulates visual cortical
plasticity. Neuron 2007, 53:747-759.
The authors identify ERK/MAPK-dependent regulation of histone modifications
as a new mechanism underlying the expression of ocular
dominance plasticity
38. Hensch TK: Critical period mechanisms in developing visual
cortex. Curr Top Dev Biol 2005, 69:215-237.
39. Quarles RH: Myelin sheaths: glycoproteins involved in their
formation, maintenance and degeneration. Cell Mol Life Sci
2002, 59:1851-1871.
40. McGee AW, Yang Y, Fischer QS, Daw NW, Strittmatter SM:
Experience-driven plasticity of visual cortex limited by myelin
and Nogo receptor. Science 2005, 309:2222-2226.
41. He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA,
Casaccia-Bonnefil P: The transcription factor Yin Yang 1 is
essential for oligodendrocyte progenitor differentiation.
Neuron 2007, 55:217-230.
42. Shen S, Casaccia-Bonnefil P: Post-translational modifications
of nucleosomal histones in oligodendrocyte lineage
cells in development and disease. J Mol Neurosci 2008,
35:13-22.
43. Shen S, Li J, Casaccia-Bonnefil P: Histone modifications
affect timing of oligodendrocyte progenitor differentiation
in the developing rat brain. J Cell Biol 2005, 169:
577-589.
44. Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJ,
Casaccia-Bonnefil P: Age-dependent epigenetic control of
differentiation inhibitors is critical for remyelination efficiency.
Nat Neurosci 2008, 11:1024-1034.
This paper provides mechanistic insight into how oligodendrocytes precursors
cell differentiation is epigenetically regulated during remyelination
and how these mechanisms change with aging.
45. Fass DM, Butler JE, Goodman RH: Deacetylase activity is
required for cAMP activation of a subset of CREB target
genes. J Biol Chem 2003, 278:43014-43019.
46. Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA,
Cabrera SM, McDonough CB, Brindle PK, Abel T et al.: Histone
deacetylase inhibitors enhance memory and synaptic
plasticity via CREB:CBP-dependent transcriptional activation.
J Neurosci 2007, 27:6128-6140.
47. Weaver IC, Meaney MJ, Szyf M: Maternal care effects on the
hippocampal transcriptome and anxiety-mediated behaviors
in the offspring that are reversible in adulthood. Proc Natl Acad
Sci U S A 2006, 103:3480-3485.